Transcending Nature: CRISPR/Cas

Fig1.
This post is a translation of my French post available here.
Nature has, over time, written the longest and most complex book to date: the genetic code.
Adenine, Thymine, Guanine and Cytosine form words defining our existence, who we are and our history.
This still globally misunderstood book contains in it the greatest secrets, of our history until our smallest characteristic.
The book is in continuous writing, evolving from cell replication to genetic mutations in such a way that we are all scribes of a wildly magnificent work allowing life in the form we know.
As scribes, we only unwittingly write this code, or at least... It's not exactly true anymore.
CRISPR/Cas, genetic scissors
Above all, it is an immune defense system present in 50 to 60% of bacteria and 90% of archaea, prokaryotic unicellular microorganisms, i.e. living organisms composed of a single cell that does not include nucleus or organelles, unlike bacteria. (Wikipédia).
The system consists of two parts:
- CRISPR, for Clustered Regularly Interspaced Short Palindromic Repeats,
- A group of genes responding to the same signals: a Cas operon.
The latter operon will be responsible for the synthesis of Cas proteins.
Viruses are read, recognized and cut to be rendered inactive.
Who says lot of species with CRISPR/Cas, also says multitude of CRISPR/Cas.
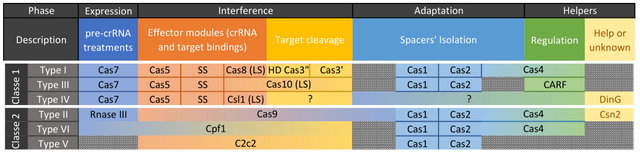
Fig2.
In our case, type II is used for its « simplicity ».
When a bacterium is infected by a virus and this infection is said abortive (without death of the organism), the adaptation enzymes Cas1 and Cas2 will include the genetic code in a hereditary database named CRISPR-array.
Each piece of virus DNA - 20 nucleotides spacers - is separated by an always identical code that can be palindromic (which can be read in both directions) although this is not the case here for CRISPR/Cas91,2,3.
We could then compare this array to a telegram encoded like this in DNA:
Virus1. Stop. Virus2. Stop. Virus3. Stop.
Later, if the same phage infects this bacterium or its progeny, the interference enzymes (Cas9 here) will come read the DNA of the phage and if it corresponds to an array's value, it cuts it up and makes it inactive4,5.
It should be noted that a new variant of Cas, named CasRx allows actions on RNA and no longer on DNA. CasRx will not be discussed here.
Technical details
At the end of the CRISPR-array sequence, is a region called Leader Sequence. It allows transcribing this array into RNA.
Via RNA expression, phagic sequences will be expressed in a long RNA sequence called pre-crRNA. This will then be recognized by a Cas protein (CRISPR-associated protein) synthesized by the operon: an endoribonuclease.
This endoribonuclease (also called RNase) will recognize each repetitions called inter-spacers, cleaving the RNA at each spacer to produce small fragments of RNA (crRNA) that will associate with a Cas protein (endonuclease Cas9).
This Cas complex will then "scan" the DNA of the phage until the RNA fragment binds to its DNA through basic complementarity.
The enzyme then produces a double strand break (breakage of the two DNA strands) via the HNH Nuclease Domain (Cyan on the picture below) and the RuvC Nuclease Domain (grey).
In nature, you need another RNA in addition to the crRNA: the tracrRNA.
The laboratories of Jennifer DOUDNA and Emmanuelle CHARPENTIER have worked together to combine these two RNAs into one: sgRNA (single guide RNA)1,2. The operation currently requires only one RNA instead of two.
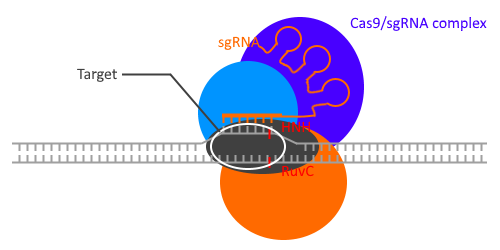
Fig3.
Once this break is made, the organism uses a mechanism called homologous recombination divided into 3 stages:
- A pre-synaptic phase (the damaged site is recognized),
- A synaptic phase (the organism looks for a similar piece of DNA (homologous) to repair the bond),
- A post-synaptic phase (DNA restoration based on the homologous model.
This implies that by injecting a CRISPR/Cas9 complex with given homologous DNA, the homologous DNA is genetically incorporated into the organism's DNA.
Perspectives et évolutions
CRISPR/Cas9 possesses many advantages. Universal, it acts as a tool requiring only RNA where old practices required a long and costly specific protein and enzyme synthesis.
The techniques are even evolving towards simplification since it is possible to select several targets by a single sgRNA via the use of universal sequences10.
It is equally possible to modify Cas9 (in "dead" Cas9) to inhibit or promote gene transcription as reported by Vijai Singh, et al. By modifying the enzyme domains Ruv-C (D10A) and HNH (H840A), the enzyme binds to DNA but can no longer cut DNA, it will interfere with gene expression.
This is called CRISPR-Interference (or CRISPRi) for inhibition (via dCas9-KRAB for mammals) and CRISPR-activation (CRISPRa) for activation (via, for example, the multiplasmid system SAM).
In the present circumstances that humans can modify DNA, anything is possible. It will then be possible to cure genetic diseases such as cystic fibrosis, hemophilia or sickle cell disease.
CRISPR/Cas9 is also considered as possible treatment against tumour cells by reversing genetic mutations.
Without mentioning the possibility of collateral damage, the complex opens up to many ethical questions in particular regarding eugenics with the modification of human embryos.
The possibilities are so infinite the American government is concerned about a future terrorism of a another kind, which could disable certain vital functions or cause important mutations.9.
References
[1.] Conférence « The CRISPR-Cas9 revolution genome engineering: lessons learned from bacteria » par Emmanuelle CHARPENTIER.
[2.] Conférence « Genome Engineering and the Future of Human Health » par Jennifer DOUDNA.
[3.] Amitai G, Sorek R. 2016. CRISPR-Cas adaptation: insights into the mechanism of action. Nat. Rev. Microbiol. 14(2):67–76.
[4.] Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, et al. 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315(5819):1709–12.
[5.] Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV. 2006. A putative RNA-interferencebased immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol. Direct 1:7.
[6.] Jörg Renkawitz, Claudio A. Lademann et Stefan Jentsch, « Mechanisms and principles of homology search during recombination », Nature Reviews. Molecular Cell Biology, vol. 15, no 6, 1er juin 2014, p. 369–383.
[7.] Vijai Singh, et al., Exploring the potential of genome editing CRISPR-Cas9 technology, 2016.
[8.] Bikard et al., 2013; Qi et al., 2013
[9.] [https://motherboard.vice.com/en_us/article/yp3xaj/obamas-science-advisors-are-worried-about-future-crispr-terrorism](https://motherboard.vice.com/en_us/article/yp3xaj/obamas-science-advisors-are-worried-about-future-crispr-terrorism)
[10.] Toon Swings, et al., CRISPR-FRT targets shared sites in a knock-out collection for off-the-shelf genome editing, 2018.
Illustrations
Fig1. Illustration personnelle, CC BY-ND Clément POIRET. Adobe Photoshop & Illustrator.
Fig2. Illustration personnelle, CC BY-SA Clément POIRET. Microsoft Excel.
Fig3. Illustration personnelle, CC BY-SA Clément POIRET. Paint(dot)Net <3.
Going further

Banner by @nitesh9 and @rocking-dave
Thanks to SteemSTEM and FrancoSTEM for their help and support! <3